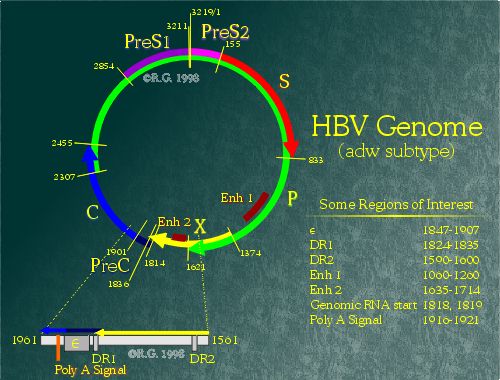
The Hepatitis B Genome
Electron microscopy gave the initial views of the hepatitis B genome. In virions, the genome appears to be circular, yet only partially double-stranded. (1) The genome is approximately 3200 nucelotides in length, but does not abide by the usual classification criteria for viruses. (2) Numbering of basepairs on the HBV genome is based on the cleavage site for the restriction enzyme EcoR1 or at homologous sites, if the EcoR1 site is absent. However, other methods of numbering are also used, based on the start codon of the core protein or on the first base of the RNA pregenome. There are at least seven major subtypes of HBV, distinguished by sequence differences in the surface antigen gene. (3)
Unlike others, HBV virions contain both DNA and RNA. Moreover, some regions of the packaged genome can be single stranded, double stranded, or even triple stranded. These peculiar features are a direct result of the HBV genome replication mechanism which will be covered later in this chapter.
There are four defined overlapping open reading frames (ORFs) in the genome which result in the transcription and expression of the seven different hepatitis B proteins. Every base pair in the HBV genome is involved in encoding at least one of the HBV protein. However, the genome also contains genetic elements which regulate levels of transcription, determine the site of polyadenylation, and even mark a specific transcript for encapsidation into the nucleocapsid.
The four ORFs lead to the transcription and translation of seven different HBV proteins through use of varying in-frame start codons. For example, the small hepatitis B surface protein is generated when a ribosome begins translation at the ATG at position 155 of the adw genome. The middle hepatitis B surface protein is generated when a ribosome begins at an upstream ATG at position 3211, resulting in the addition of 55 amino acids onto the 5' end of the protein. ORF P occupies the majority of the genome and encodes for the hepatitis B polymerase protein. ORF S encodes the three surface proteins. (4) ORF C encodes both the hepatitis e and core protein. (5) ORF X encodes the hepatitis B X protein. (6), (7)
Promoters and Signal Regions
The HBV genome contains many important promoter and signal regions necessary for viral replication to occur.
The four ORFs transcription are controlled by four promoter elements (preS1, preS2, core and X), and two enhancer elements (Enh I and Enh II). All HBV transcripts share a common adenylation signal located in the region spanning 1916-1921 in the genome. Resulting transcripts range from 3.5 nucleotides to 0.9 nucleotides in length. Due to the location of the core/pregenomic promoter, the polyadenylation site is differentially utilized. (8) The polyadenylation site is a hexanucleotide sequence (TATAAA) as opposed to the canonical eukaryotic polyadenylation signal sequence (AATAAA). The TATAAA is known to work inefficiently (9), suitable for differential use by HBV.
The core/pregenome promoter controls the transcription of multiple RNAs which have 5' heterogeneity. These include the core antigen, e antigen, polymerase, and pgRNA. These transcripts are also regulated by both viral enhancer elements and a negative regulatory element (NRE). The core/pregenome promoter has yet to be clearly defined, but has been mapped loosely in a region spanning nucleotides 1591 to 1851. (10), (11) Positive regulation of this region involves many transacting cellular factors. CCAAT/enhancer binding protein (C/EBP), (12), HNF-3, (13) HNF-4, (14) and an ubiquitous cellular factor, Sp1 (15), appear to assist in activation. Studies have also indicated that members of the nuclear receptor family may be involved in downregulation of the core/pregenome promoter. (16), (17) The polymerase coding region does not appear to have a promoter element directly upstream. Its expression is believed to be the result of ribosome scanning of the pgRNA transcript.
ORF S contains three in-frame start sites which direct synthesis of the three distinct hepatitis B surface proteins. (18), (19), (20) There are two promoter regions which control the expression of these proteins, namely the preS1 promoter and preS2 promoter. The preS1 promoter controls transcription of a single 2.4kb RNA molecule which includes the entire ORF S region. However, the preS2 promoter controls transcription of a family of transcripts of 2.1kb in length. The preS2 promoters has been shown to be stronger than the preS1 promoter. (21), (22), (23) This results in more middle and small surface proteins being expressed as compared to the large surface protein. It has been shown that overexpression of the large surface protein results in retention of surface proteins in the endoplasmic reticulum. (24), (25), (26) Transcription factors such as Oct-1 (27), HNF-1 and HNF-3 (28), (29), (30) appear to be involved in the activation of the preS1 promoter. HBV enhancer elements also appear to be involved in preS1 promoter upregulation. (31), (32) Downregulation of the preS1 promoter appears to be dependent on a region within the preS2 promoter (nucleotides 3160 to 3221). (33) However, the mechanism is poorly understood. The preS2 promoter appears to be upregulated by a CCAAT element which is also involved in the downregulation of the preS1 promoter. NF-Y, the CCAAT-binding factor, appears to be involved in the preS2 activation (34) as well as both HBV enhancers. (35), (36) Also, stress on the endoplasmic reticulum due to the presence of hepatitis B large surface protein appears to result in increased activation. (37)
ORF X, which encodes a 17kd protein known as the hepatitis B X Protein (HBx), has its own promoter controlling the transcription of a 0.9kb RNA. However, due to the proximity of the X promoter to enhancer I region, the precise borders remain controversial.The promoter is believed to lie within the region spanning nucleotides 1230 to 1376. (38), (39)
The enhancer I element spans a region from nucleotides 970-1240. (40), (41) Many transcription factors are believed to interact with this region of the HBV genome. These molecules include HNF-3, (42) HNF-4, (43) EF-C, (44) and NF-1, to name a few. Both enhancer I and enhancer II can activate heterologous promoters despite their position or orientation. (45), (46) Deletion of either enhancer region results in a strong reduction of viral transcripts. (47)
The last region which will be mentioned in this section is known as the epsilon-stem loop.(-stem loop) The region spans nucleotides 1847-1907 in the HBV genome and plays a key role in HBV DNA encapsidation. The location of the stem-loop was determined by fusing heterologous genes to various regions of the HBV genome and by looking for encapsidation of these foreign genes. (48), (49), (50) Despite the terminal redundancy of the pgRNA, only the 5' -stem loop retains functionality. (51) Because of this, only pgRNAs are encasidated despite the fact that all HBV transcripts have the -stem loop coding region at their 3' ends. Sequence analysis of the -stem loop shows a series of inverted repeats that are predicted to fold into a three-dimensional stem-loop structure. This stem-loop is conserved among all hepadnaviruses despite differences in the primary sequence. (52) It is believed that the polymerase protein recognizes and interacts directly with the stem-loop structure. (53) Interaction of the hepatitis B polymerase and the -stem-loop initiates both encapsidation as well as reverse transcription of the HBV pgRNA.
The genomic arrangement of the hepatits B virus family makes it unique among viruses. The unusual nature of the packaged genome gives an indication that the method of replication employed by HBV is not that of conserved DNA replication. We shall look more closely at the mechanisms members of the hepadnaviridae use when replicating in the following section.
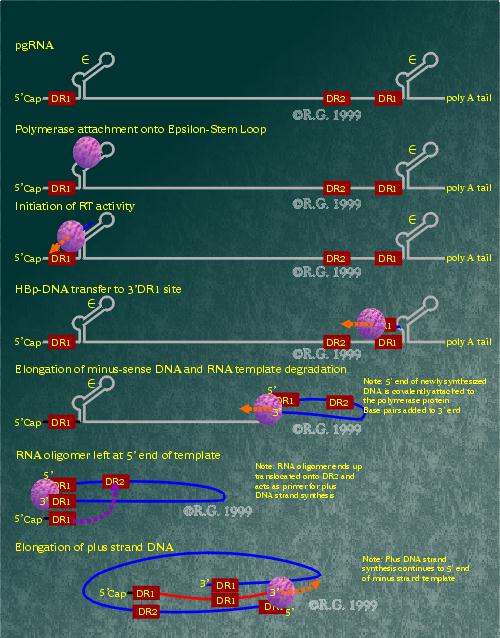
Multiplication of the HBV genome occurs within the nucleus of an infected cell. RNA polymerase II transcribes the circular HBV DNA into greater-than-full length mRNA. Since the mRNA is longer than the actual cccDNA, redundant ends are formed. Once produced, the genomic RNA exits the nucleus and enters the cytoplasm whereupon it can be translated to generate the HBV polymerase, core and e proteins.
The pregenomic RNA packaging into core particles is triggered by the binding of the HBV polymerase to the 5' epsilon stem-loop. RNA encapsidation is believed to occur as soon as binding occurs. The HBV polymerase also appears to require associated core protein in order to function. The HBV polymerase initiates reverse transcription from the 5' epsilon stem-loop three to four base pairs, at which point, the polymerase and attached nascent DNA are transferred to the 3' copy of the DR1 region. Once there, the (-)DNA is extended by the HBV polymerase while the RNA template is degraded by the HBV polymerase RNAse H activity. When the HBV polymerase reaches the 5' end, a small stretch of RNA is left undigested by the RNAse H activity. This segment of RNA is comprised of a small sequence just upstream, including the DR1 region. The RNA oligomer is then translocated and annealed to the DR2 region at the 5' end of the (-)DNA. It is used as a primer for the (+)DNA synthesis which is also generated by the HBV polymerase. It appears that the reverse transcription as well as plus strand synthesis may occur in the completed core particle. Because of this, the finite amount of free nucleotides within the core particle may be the reason that the (+)DNA strand of the HBV genome varies in length in completed virions.
Types
of HBV Genomes
The following is a list of the major types of HBV genomes
found in the human population:
-
1993: Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen.
-
1995: Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene.
-
1998: Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela.
6 Genotypes (A,B,C,D,E,F) [PMID: 8336122]
-
Group A - Orig - N. Europe - Sub-Saharan Africa
-
Group B - Confined to - Eastern Asia (China)
-
Group C - Far East (Japan)
-
Group D - Mediterranean - Near, Mid East, South Asia
-
Group E - W. Sub-Saharan Africa, south to Angola
-
Group F - New World - Brazil, N. + S. America
121 Strains Exist as Quasispecies
There are four serotypes which are based on subtypes of the hepatitis B surface antigen (HBsAg). These are defined by two mutually exclusive determinant pairs d/y and w/r with a common determinant 'a'. These subtypes are adw, ayw, adr, and ayr.
Four genomic groups of HBV were later referred as genotypes designated with A-D. Sequencing of the S-gene of HBV is the molecular basis for the assessment of the serological variations of HBsAg within the major four subtypes. Two new genotypes of HBV are designated with E and F. The F genotype diverges from other HBV genomes sequenced by 14%. So far, it is the most divergent HBV genome. Worldwide molecular epidemiology of HBV is based on the variability of the S-gene. The E and F strains appear to originate from aboriginal populations of Africa in the New World.
References:
1.
Robinson et al. 1974. DNA of a human hepatitis B virus candidate.
J. Virol.; 14: 384-391.
2. Francki, R.I.B. et. al. 1991. Classification and nomenclature of viruses. Springer, Vienna.
3. Raney, A.K. and McLachlan, A. 1991. In Molecular Biology of the Hepatitis B Virus ed. A. McLachlan, pp. 1-38. CRC Press, Boca Raton, Florida.
4. Heermann K.H. et al. 1984. Large surface proteins of hepatitis B virus containng the pre-S sequence. J. Virol; 52: 396-402.
5. Ou, J.H. et al. 1986. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membrances and causes the secretion of the e antigen. Proc. Natl. Acad. Sci USA; 83: 1578-1582.
6. Kwee, L. et al. 1992. Alternate translation initiation on hepatitis B virus X mRNA produced multiple polypeptides that differentially transactive class II and III promoters. J. Virol.; 66: 4382-4389.
7. Tiollais, P. et al. 1985. Nature; 317: 489-
8. Russnak, R. and Ganem, D. 1990. Genes and Dev.;4: 764-
9. Sheets, M. et al. 1990. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res; 18: 5799-5805.
10. Lopez-Cabrera et al. 1990. Proc. Natl. Acad. Sci. USA; 87: 5069-
11. Yuh et al. 1992. J. Virol; 66: 4073-
12. Lopez-Cabrera et al. 1990. Proc. Natl. Acad. Sci. USA; 87: 5069-
13. Li, M. et al. 1995. Virology; 214: 371-
14. Guo et al., 1993. Mol. Cell. Bio; 13: 443-
15. Zhang, P. et al. 1993. J. Virol; 1472-
16. Raney, A.K. et al. 1997. J. Virol; 71: 1058-
17. Yu, X. and Mertz, J.E. 1997. J. Viro.; 71: 9366-
18. Eble, B.E., Lingappa, V.R. and Ganem, D. 1986. Hepatitis B Surface Antigen: An Unusual Secreted Protein Initially Synthesized as a Transmembrane Polypeptide. Mol and Cell Biology; 6: 1454-1463.
19. Heermann, K.H., Goldmann, U., Schwartz, W. Seyffarth, T., Baumgarten, H. and Gerlich, W.H. 1984. Large Surface Proteins of Hepatitis B Virus Containing the Pre-S Sequence. J Virol; 52: 396-402.
20. Heermann, K.H. and Gerlich, W.S. 1991. Surface Proteins of Hepatitis B Viruses. In: McLachlan, A. (ed.) Molecular Biology of the Hepatitis B Viruses. CRC Press, Boca Raton, pp. 109-144.
21. Schaller, H. and Fischer, M. 1991. In Current Topics in Microbiology and Immunology, ed. W.S.Mason and C.Seeger, pp. 21-39. Springer-Verlag, Berlin-Heidleberg, Germany.
22. Shaul, Y. 1991. In Molecular Biology of Hepatitis B Virus. ed. A. McLachlan, pp. 193-211. CRC Press, Boca Raton, Florida.
23. Yen, T.S.B. 1993. Seminars in Virology; 4: 33-
24. Persing, D.H. 1986. Science; 234: 1388-
25. Chisari, F.V. et al. J. Virol; 60:880-
26. Strandring, D.N. et al., Proc. Natl. Acad. Sci. USA.; 83: 9338-
27. Zhou, D.X. and Yen, T.S.B. 1991, Mol. Cell. Biol; 11: 1353-
28. Courtois, G. et al., Proc. Natl. Acad. Sci. USA; 85: 7937-
29. Zhou, D.X. and Yen, T.S.B. 1991, Mol. Cell. Biol; 11: 1353-
30. Raney, A.K. et al. 1995 J. Virol.; 69: 3265-
31. Yuh, C.H. and Ting, L.P.J., J. Virol.;64: 4281-
32. Su, H. and Yee, J.K. Proc. Natl. Acad. Sci. USA; 89: 2708-
33. Bulla, G.A. and Siddiqui, A. 1989, Virology; 170: 251-
34. Lu, C.C. and Yen. T.S.B. 1996. Virology; 225: 387-
35. Bulla, G.A. and Siddiqui, A. 1988, J.Virol.; 62: 1437-
36. Raney, A.K. et al. 1989, J. Virol.; 63: 3919-
37. Xu, Z., Jensen, G. and Yen, T.S. 1997. Activation of Hepatitis B Virus S Promoter by the Viral Large Surface Protein via Induction of Stress in the Endoplasmic Reticulum. J Virol; 71(10): 7387-7392.
38. Treinin, M. and Laub, O. 1987. Mol. Cell. Bio.; 7: 545-
39. Siddiqui, A. et al., 1987. Proc. Natl. Acad. Sci. USA; 84: 2513-
40. Shaul, Y. et al., EMBO K.; 4: 427-
41. Tognoni, A. et al., 1985. Nuc. Acid. Res.; 13: 7457-
42. Chen, M. et al. 1994. Virology; 205:127-
43. Garcia, A.D. et al. 1993, J. Virol.; 67: 3940-
44. Ostapchuk, P. et al. 1989, Mol. Cell. Bio.; 9: 2787-
45. Yee, J. 1989. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science; 246: 658-670.
46. Yen, T.S.B. 1993. Regulation of hepatitis B virus gene expression. Semin. Virol.; 4: 33-42.
47. Lopez-Cabrera, M. et al. 1990. Transcriptional factor C/EBP binds to and transactivated the enhancer II element of the hepatitis B virus. Virology; 183: 825-829.
48. Chiang, P-W et al. 1992. Characterization of a cis element required for packaging and replication of the human hepatitis B virus. Virology; 186: 701-711.
49. Junker-Niepmann, M. et al, 1990. A short cis-acting sequences is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of feoreign RNA. EMBO J.; 9: 3389-3396.
50. Pollack, J et al., 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol; 67: 3254-3263.
51. Hirsch, R. et al., 1991. Cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol.; 65: 3309-3316.
52. Junker-Niepmann, M et al., 1990. A short cis-acting sequence is required for hepatitis B virus pregome encapsidation and sufficient for packaging of foreign RNA. EMBO J; 9: 3389-3396.
53. Pollack, J et al. 1994. Site-specific RNA bindding by a hepatitis B virus reverse transcriptase initiates two reactions: RNA packaging and DNA synthesis. J. Virol.; 68: 5579-5587.
