Hepatitis B Core and e Proteins
The core ORF encodes two sequence related yet functionally distinct proteins: the hepatitis B core protein and the hepatitis B e protein.
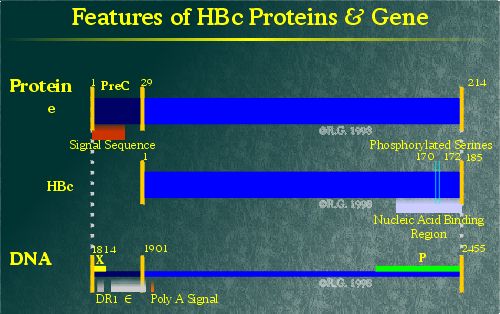
The hepatitis B core protein (HBc) is the major component of the nucleocapsid shell packaging the HBV genome. This 185 amino acid protein is expressed in the cytoplasm of infected cells. Sequence analysis indicates that HBc is predominantly composed of hydrophilic and charged amino acids. This protein remains non-glycosylated and is not modified through the addition of lipids.
The region of HBc associated with nucleocapsid assembly lies within the first 149 amino acids. (1) More recently, the crystal structure of the nucleocapsid has been resolved, giving more insight into the structure of the HBc protein. (2) HBc is largely a helical protein with five alpha-helices. This is unusual in that most nucleocapsid proteins which form icosahedral shells typically fold into barrel structures such as those seen in the HIV-1 capsid. (3), (4) A long alpha-helical hairpin dominates the folding pattern of HBc, consisting of helices 2 and 3. A hydrophobic core makes up the base of the protein.
HBc dimers, required for the formation of the nucleocapsid, associate via their alpha-helical hairpins which form a four-helix bundle. (5) A disulfide bridge on the dimer interface between the Cys-61 residues forms to stabilize the association. (6), (7) However, this residue is not critical as its deletion does not impair dimer formation. (8), (9), (10) Dimer associations to produce the nucleocapsid shell appear to involve residues Tyr-132, Arg-127, Pro-129 and Ile-139. (11)
The C-terminal end of the protein contains four arginine clusters which appear to be involved in nucleotide packaging. (12), (13) HBc can also be phosphorylated (14), (15) on serine residues at positions 157, 164 and 172. Phosphorylation at series 164 and 172 impairs pgRNA packaging. (16), (17)
HBc is able to form particles with outward pointing spikes even if a foreign peptide sequence is spliced into the HBc coding region. As such, HBc can be used for displaying foreign protein domains on the surface of HBV nucleocapsids. (18)
Unlike the core protein, the hepatitis B e antigen has a different fate. The e antigen is so named due to its "early" appearance during an acute HBV infection. HBe is expressed by the translation of sequences upstream of the HBc ORF known as the pre-C sequence. The pre-C sequence encodes a hydrophobic transmembrane domain, resulting in the translation/translocation of HBe into the lumen of the ER. (19) This fundamental change in the location of protein expression alters the antigenicity of HBe such that it does not share antigenic homology with HBc, despite having nearly identical amino acid sequences.
In the ER, 19 of the 29 residues of the pre-C region are removed by a signal peptidase. The remaining pre-C residues prevent HBe from forming into core particles. (20) Another region in the arginine rich domain is cleaved from HBe in the Golgi complex. (21)
The purpose of producing this secretory form of HBc is not well understood. One theory is that high levels of HBe may somehow suppress the immune system from eliminating HBV-producing cells. High levels of HBe are often found in the serum of highly varaemic HBV carriers. It has also been found that woodchucks do not undergo persistent WHV infection if a HBe-minus variant is used for infection. (22)
References
1.
Gallina, A., Bonelli, F., Zentilin, L, Rindi, G., Muttini, M. and
Milanesi, G. 1989. A Recombinant Hepatitis B Core Antigen Polypeptide
with the Protamine-like Domain Deleted Self-Assembles into Capsid Particles
But Fails to Bind Nucleic Acids. J.Virol.; 63: 4645-4652.
2. Wynne et al., 1999. The crystal structure of the human hepatitis B capsid. Mol. Cell. Bio.
3. Momany et al., 1996.
4. Gamble et al., 1997.
5. Wynne et al., 1999. The crystal structure of the human hepatitis B capsid. Mol. Cell. Bio.
6. Nassal, M. 1992. The Arginine-Rich Domain of the Hepatitis B Virus Core Protein is Required for Pregenome Encapsidation and Productive Viral Positive-Strand DNA Synthesis But Not for Virus Assembly. J. Virol.; 66: 4107-4116.
7. Zheng et al., 1992.
8. Nassal et al., 1992.
9. Zheng et al., 1992.
10. Zlotnick et al., 1996.
11. Wynne et al., 1999. The crystal structure of the human hepatitis B capsid. Mol. Cell. Bio.
12. Hatton, T., Zhou, S. and Standring, D.N. 1992. RNA and DNA-Binding Activities in Hepatitis B Virus Capsid Protein A Model for their Roles in Viral Replication. J. Virol.; 66: 5232-5241.
13. Nassal et al., 1992.
14. Roosinck, M.J. and Siddiqui, A. 1987. In vivo Phosphorylation and Protein Analysis of Hepatitis B Virus Core Antigen. J. Virol.; 61: 955-961.
15. Lan et al., 1999. Three major phosphorylation sites of hepatitis B core protein in viral replication.
16. Lan et al., 1999.
17. Machida, A., Ohnuma, H. and Tsuda, F. 1991. Phosphorylation in the Carboxyterminal Domain of the Capsid Protein of Hepatitis B Virus: Evaluation with a Monoclonal Antibody. J. Virol.; 12: 1017-1021.
18. Kratz et al., 1999. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA.
19. Bruss, V. and Gerlich, W.H. 1988. Formation of Transmembranous Hepatitis B e-antigen by Cotranslational in vitro Processing of the Viral Precore Protein. Virology; 163: 268-275.
20. Wasenauer, G., Kock, J. and Schlicht, H.J. 1992. A Cysteine and a Hydrophobic Sequence in the Noncleaved Portion of the Pre-C Leader Peptide Determine the Biophysical Properties of the Secretory Core Protein (HBe protein) of Human Hepatitis B Virus. J. Virol.; 66: 5338-5346.
21. Standring, D.N., Ou, J.H., Masiarz, F.R. and Rutter, W.J. 1988. A Signal Peptide Encoded within the Precore Region of Hepatitis B Virus Directs the Secretion of a Heterogeneous Population of e Antigens in Xenopus Oocytes. Proc Natl Acad Sci USA; 85: 8405-8409.
22. Chen, H.S., Kew, M.C, Hornbucke, W.E. et al. 1992. The Precore Gene of the Woodchuck Hepatitis Virus Genome is not Essential for Viral Replication in the Natural Host. J. Virol.; 66: 5682-5684.
